Pipes
Joule pipes provide a protocol independent interface to data
streams. This decouples module design from pipeline
implementation. There are three different pipe implementations,
joule.LocalPipe, joule.InputPipe, and joule.OutputPipe all of which derive
from the abstract base class
joule.Pipe. LocalPipe's are intended for intra-module communication (see Composite Modules).
It is bidirectional meaning it supports both read and write operations. InputPipe's and OutputPipe are unidirectional
supporting only read and write respectively. These are intended for inter-module communication with
one module's OutputPipe connected to another's InputPipe. The figure below
illustrates how pipes move stream data between modules.
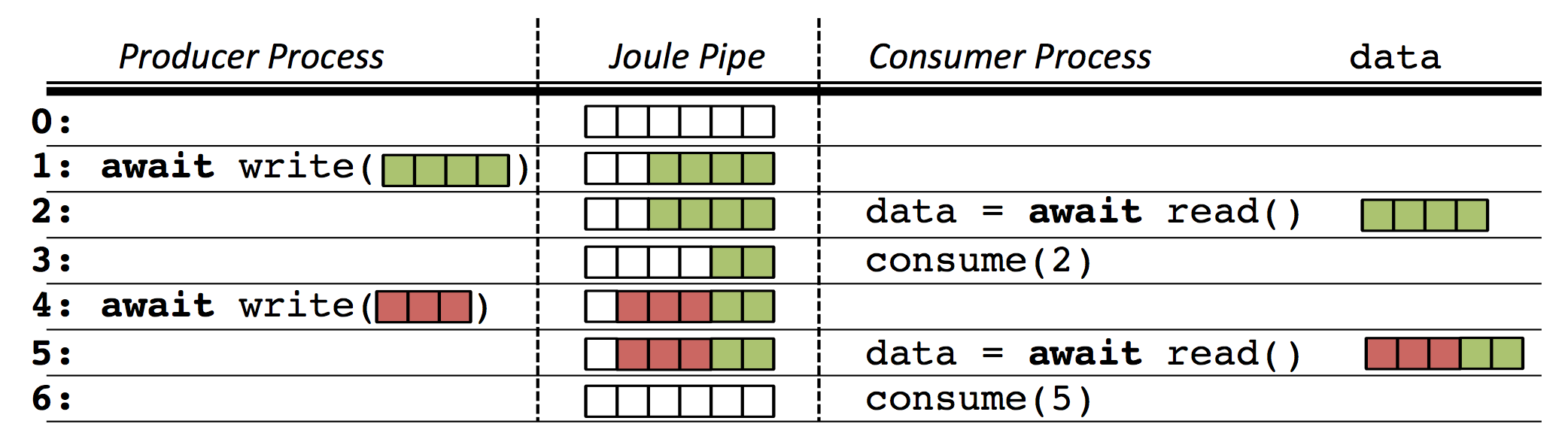
In the figure the pipe is initially empty at Time 0. At Time 1, the producer adds four rows of data which the consumer reads at Time 2. The consumer only consumes two rows so the last two rows of data remain in the pipe. At Time 4 the producer writes three more rows of data which are appended to the two unconsumed rows. The next call to read returns all five rows of data. This time the consumer consumes all the data and the pipe is empty. Conceptually the pipe is a buffer between the producer and consumer, however it is important to note that these are independent processes and may not even be running on the same machine.
Note
When designing modules care must be taken to ensure that they execute fast enough to handle streaming data. If a module’s memory usage increases over time this indicates the module cannot keep up with the input and the Joule Pipe buffers are accumulating data.
Intervals
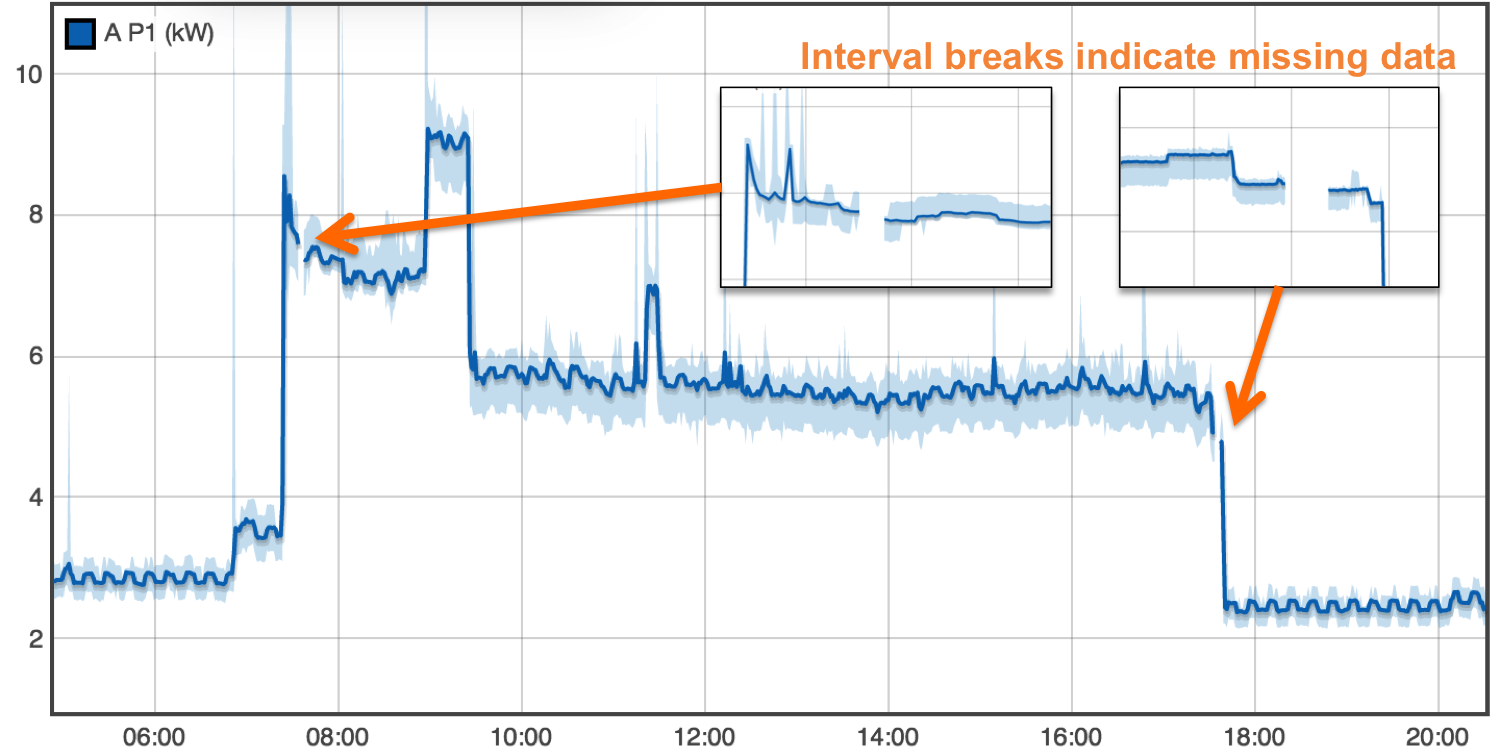
Streams are divided into intervals of continuous time series data. The first write to a pipe starts a new stream interval. Subsequent writes append data to this interval. This indicates to data consumers that there are no missing samples in the stream. To indicate missing data the producer closes the interval. A new interval is started on the next write. The plot below shows a stream with three seperate intervals indicating two regions of missing data.
- Data Producers
The code snippet below shows how a data producer indicates missing samples using intervals. In normal operation the sensor output is a single continuous stream interval. If the sensor has an error the exception handler closes the current interval and logs the event. See Intermittent Reader for a complete example.
while True:
try:
data = sensor.read()
await pipe.write(data)
except SensorException:
log.error("sensor error!")
await pipe.close_interval()
- Data Consumers
The code snippet below shows how a data consumer detects interval breaks in a stream. At an interval boundary read will return data up to the end of the current interval and set the end_of_interval flag. The next call to read will return data from the new interval and clear the flag. Any unconsumed data will be returned with the next read even though it is from a previous interval. Therefore it is best practice to completely consume data on an interval break and reset any buffers to their initial state as suggested with the fir_filter logic below. See Offset Filter for a complete example on handling interval boundaries.
while True:
data = await pipe.read()
pipe.consume(len(data))
output = fir_filter.run(data)
if(pipe.end_of_interval):
fir_filter.reset()
In general filters should propagate input interval breaks to their outputs. In other words unless a filter can restore missing input data, it should have at least as much missing output data. The snippet below shows the logic for propagating interval breaks.
data = await input_pipe.read()
input_pipe.consume(len(data))
await output_pipe.write(data)
if data.end_of_interval:
await output_pipe.close_interval()
Caching
By default a call to write will immediately send the data to the transport layer (OS pipe, network socket, etc). In order to reduce the overhead associated with the transport layer data should be batched. Data may be batched manually (see high_bandwidth_reader) or you may use the pipe cache to perform data batching. When the cache is enabled the transport layer will only be executed when the specified number of rows have been written. This eliminates the performance penalty of frequent short writes. Timestamps should be linearly interpolated for high bandwidth data rather than individually timestamped. The cache should be sized to execute writes at about 1 Hz.
# sensor produces data at 1KHz
time = time.now()
pipe.enable_cache(1000)
while(1):
data_point = sensor.read()
ts += 1000 #1ms = 1000us
await pipe.write([[ts,data_point]])
Subscriptions
A single input can be copied to multiple outputs using pipe subscriptions. Pipes that produce output (OutputPipe or LocalPipe) A LocalPipe can subscribe to input end of a LocalPipe can be subscribed to either an OutputPipe or the output end of another LocalPipe.
Note
Pipe subscriptions are only necessary for creating data paths within Composite Modules. Joule manages inter-module data paths automatically
p1.subscribe(p2)
await p1.write([1,2,3])
await p1.read() # 1,2,3
p1.consume(len(data))
# p2 still has the data
p2.read() # 1,2,3
p2.consume(2)
await p1.write([4,5,6])
p1.read() # 4,5,6
p2.read() # 3,4,5,6
Reference
- class joule.Pipe(name=None, direction=None, module=None, stream=None, layout=None)[source]
This encapsulates streams and connects to modules. Some more infos2
Note
There are many different kinds of pipes
- async close()[source]
Close the pipe. This also closes any subscribers. If
close_cbis defined it will be executed before the subscribers are closed.
- async close_interval()[source]
Signal a break in the data stream. This should be used to indicate missing data. Data returned from
read()will be chunked by interval boundaries.
- close_interval_nowait()[source]
Signal a break in the data stream. This will dumped cached data and should generally not be used. Instead use the coroutine
close_interval().
- consume(num_rows)[source]
Flush data from the read buffer. The next call to
read()will return any unflushed data followed by new incoming data.- Parameters:
num_rows -- number of rows to flush from the read buffer
- consume_all()[source]
Flush all data from the read buffer. The next call to
read()will only return new incoming data.
- enable_cache(lines: int)[source]
Turn on caching for pipe writes. Data is only transmitted once the cache is full. This improves system performance especially if
write()is called rapidly with short arrays. Once enabled, caching cannot be disabled.- Parameters:
lines -- cache size
- async flush_cache()[source]
Force a pipe flush even if the cache is not full. Raises an error if caching is not enabled.
- async read(flatten=False) ndarray[source]
Read stream data. By default this method returns a structured array with
timestampanddatafields. This method is a coroutine.- Parameters:
flatten -- return an unstructured array (flat 2D matrix) with timestamps in the first column
- Returns:
numpy.ndarray
>>> data = await pipe.read() [1, 2, 3] >>> data = await pipe.read(flatten=True) # the same data is returned again [1,2,3] >>> pipe.consume(len(data)) # next call to read will return only new data
- async read_all(flatten=False, maxrows=100000, error_on_overflow=False) ndarray[source]
Read stream data. By default this method returns a structured array with
timestampanddatafields. The pipe is automatically closed. This method is a coroutine.- Parameters:
flatten -- return an unstructured array (flat 2D matrix) with timestamps in the first column
maxrows -- the maximum number of rows to read from the pipe
error_on_overflow -- raise a PipeError exception if pipe is not empty after reading maxrows
- Returns:
numpy.ndarray
>>> data = await pipe.read_all(flatten=True) [1, 2, 3]
- reread_last()[source]
The next read will return only unconsumed data from the previous read and no new data from the source. The end_of_interval flag is maintained.
- async write(data)[source]
Write timestamped data to the pipe. Timestamps must be monotonically increasing and should not overlap with existing stream data in the database. This method is a coroutine.
- Parameters:
data (numpy.ndarray) -- May be a structured array with
timestampanddatafieldscolumn. (or an unstructured array with timestamps in the first) --
>>> await pipe.write([[1000, 2, 3],[1001, 3, 4]])
- class joule.InputPipe(name=None, stream=None, layout=None, reader=None, reader_factory=None, close_cb=None, buffer_size=10000)[source]
- BUFFER_SIZE
The StreamReader.read coroutine hangs even if the write side of the pipe is closed so the call is wrapped in a wait_for
- Type:
Note
- async close()[source]
Close the pipe. This also closes any subscribers. If
close_cbis defined it will be executed before the subscribers are closed.
- consume(num_rows)[source]
Flush data from the read buffer. The next call to
read()will return any unflushed data followed by new incoming data.- Parameters:
num_rows -- number of rows to flush from the read buffer
- consume_all()[source]
Flush all data from the read buffer. The next call to
read()will only return new incoming data.
- async read(flatten=False) ndarray[source]
Read stream data. By default this method returns a structured array with
timestampanddatafields. This method is a coroutine.- Parameters:
flatten -- return an unstructured array (flat 2D matrix) with timestamps in the first column
- Returns:
numpy.ndarray
>>> data = await pipe.read() [1, 2, 3] >>> data = await pipe.read(flatten=True) # the same data is returned again [1,2,3] >>> pipe.consume(len(data)) # next call to read will return only new data
- class joule.OutputPipe(name=None, stream=None, layout=None, close_cb=None, writer: StreamWriter | None = None, writer_factory=None)[source]
- async close()[source]
Close the pipe. This also closes any subscribers. If
close_cbis defined it will be executed before the subscribers are closed.
- async close_interval()[source]
Signal a break in the data stream. This should be used to indicate missing data. Data returned from
read()will be chunked by interval boundaries.
- close_interval_nowait()[source]
Signal a break in the data stream. This will dumped cached data and should generally not be used. Instead use the coroutine
close_interval().
- enable_cache(lines: int)[source]
Turn on caching for pipe writes. Data is only transmitted once the cache is full. This improves system performance especially if
write()is called rapidly with short arrays. Once enabled, caching cannot be disabled.- Parameters:
lines -- cache size
- async flush_cache()[source]
Force a pipe flush even if the cache is not full. Raises an error if caching is not enabled.
- async write(data)[source]
Write timestamped data to the pipe. Timestamps must be monotonically increasing and should not overlap with existing stream data in the database. This method is a coroutine.
- Parameters:
data (numpy.ndarray) -- May be a structured array with
timestampanddatafieldscolumn. (or an unstructured array with timestamps in the first) --
>>> await pipe.write([[1000, 2, 3],[1001, 3, 4]])
- class joule.LocalPipe(layout: str, name: str | None = None, close_cb=None, debug: bool = False, stream=None, write_limit=0)[source]
Pipe for intra-module communication.
- Parameters:
layout --
datatype_width, for examplefloat32_3for a three element stream must. See DataStream.layout- Keyword Arguments:
name -- useful for debugging with multiple pipes
close_cb -- callback coroutine executed when pipe closes
debug -- enable to log pipe usage events
- async close()[source]
Close the pipe. This also closes any subscribers. If
close_cbis defined it will be executed before the subscribers are closed.
- async close_interval()[source]
Signal a break in the data stream. This should be used to indicate missing data. Data returned from
read()will be chunked by interval boundaries.
- close_interval_nowait()[source]
Same as close_interval but this is not a coroutine. This should only be used for unit testing
- close_nowait()[source]
Same as close but this is not a coroutine. This should only be used for unit testing
- consume(num_rows)[source]
Flush data from the read buffer. The next call to
read()will return any unflushed data followed by new incoming data.- Parameters:
num_rows -- number of rows to flush from the read buffer
- consume_all()[source]
Flush all data from the read buffer. The next call to
read()will only return new incoming data.
- enable_cache(lines: int)[source]
Turn on caching for pipe writes. Data is only transmitted once the cache is full. This improves system performance especially if
write()is called rapidly with short arrays. Once enabled, caching cannot be disabled.- Parameters:
lines -- cache size
- async flush_cache()[source]
Force a pipe flush even if the cache is not full. Raises an error if caching is not enabled.
- async read(flatten=False) ndarray[source]
Read stream data. By default this method returns a structured array with
timestampanddatafields. This method is a coroutine.- Parameters:
flatten -- return an unstructured array (flat 2D matrix) with timestamps in the first column
- Returns:
numpy.ndarray
>>> data = await pipe.read() [1, 2, 3] >>> data = await pipe.read(flatten=True) # the same data is returned again [1,2,3] >>> pipe.consume(len(data)) # next call to read will return only new data
- read_nowait(flatten=False)[source]
Same as read but this is not a coroutine. This should only be used for unit testing.
- Parameters:
flatten --
- Returns:
numpy.ndarray
>>> data = pipe.read_nowait() [1, 2, 3]
- reread_last()[source]
The next read will return only unconsumed data from the previous read and no new data from the source. The end_of_interval flag is maintained.
- async write(data: ndarray)[source]
Write timestamped data to the pipe. Timestamps must be monotonically increasing and should not overlap with existing stream data in the database. This method is a coroutine.
- Parameters:
data (numpy.ndarray) -- May be a structured array with
timestampanddatafieldscolumn. (or an unstructured array with timestamps in the first) --
>>> await pipe.write([[1000, 2, 3],[1001, 3, 4]])